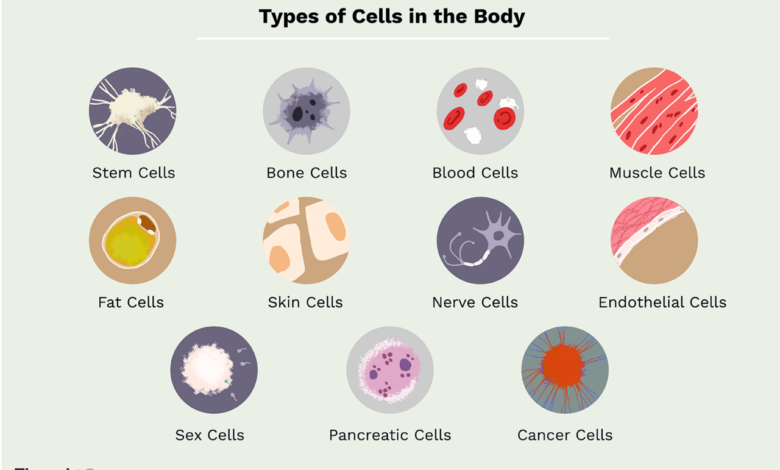
Scientists Are Building a Cell Body Catalogue
Scientists are building a catalogue of every type of cell in our bodies – a mind-blowing project, right? Imagine having a complete map of every single cell in the human body, detailing its function, location, and characteristics. This isn’t science fiction; it’s the ambitious goal of the Human Cell Atlas project. Through cutting-edge technologies like single-cell RNA sequencing, researchers are painstakingly identifying and classifying the trillions of cells that make us who we are.
The implications are vast, promising breakthroughs in disease diagnosis, personalized medicine, and our fundamental understanding of human biology.
This incredible undertaking involves international collaboration, advanced technology, and a huge amount of data analysis. Think of it as creating a detailed instruction manual for the human body, down to the cellular level. This level of understanding could revolutionize how we approach healthcare, paving the way for more effective treatments and earlier diagnoses of diseases. It’s a monumental task, but the potential rewards are simply immense.
Cell Type Classification and Identification
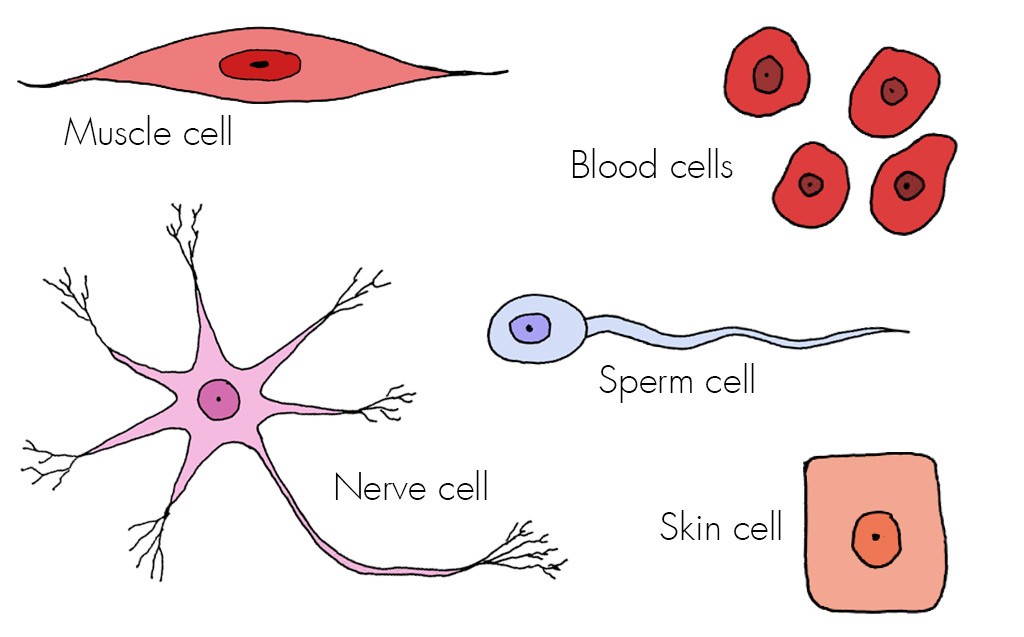
The human body is a breathtakingly complex ecosystem, composed of trillions of cells working in concert. Understanding this intricate cellular landscape requires sophisticated methods for distinguishing between the vast array of cell types. This involves not only identifying individual cells but also classifying them based on their characteristics and functions, ultimately building a comprehensive cell atlas.Cell type classification relies on a combination of techniques, each offering unique insights into cellular identity.
These methods can be broadly categorized into those that analyze morphology, those that examine protein expression, and those that investigate gene expression.
Morphological Analysis
Microscopic examination remains a cornerstone of cell identification. Different cell types exhibit distinct shapes, sizes, and internal structures. For example, neurons are easily recognizable by their long, branching axons and dendrites, while epithelial cells often form sheets with tightly connected junctions. However, morphological analysis alone is often insufficient for precise classification, especially when dealing with subtle differences between cell types.
Limitations include subjective interpretation and the inability to distinguish cells with similar morphology but distinct functions.
Protein Expression Analysis
Immunohistochemistry (IHC) and flow cytometry are powerful techniques for identifying cell types based on their protein expression profiles. IHC uses antibodies to detect specific proteins within tissue samples, providing spatial information about protein localization. Flow cytometry, on the other hand, allows for high-throughput analysis of individual cells, measuring the expression of multiple proteins simultaneously. For example, identifying immune cells often relies on the expression of specific surface markers like CD4 or CD8.
While powerful, these methods are limited by the number of proteins that can be assessed simultaneously and the potential for cross-reactivity of antibodies.
Gene Expression Analysis: Single-Cell RNA Sequencing
Single-cell RNA sequencing (scRNA-seq) represents a revolutionary advance in cell type identification. This technique allows for the analysis of the transcriptome – the complete set of RNA transcripts – of individual cells. This provides an incredibly detailed picture of gene expression, revealing subtle differences between cell types that might be missed by other methods. The process typically involves isolating individual cells, converting their mRNA into cDNA, amplifying the cDNA, and then sequencing it.
The resulting data is then used to cluster cells based on their gene expression profiles, identifying distinct cell populations. For example, scRNA-seq has been instrumental in identifying rare cell types within complex tissues like the brain and the immune system. Further analysis using bioinformatics tools allows researchers to link gene expression patterns to cell function and developmental origin.
Cell Type Identification Flowchart
The process of cell type identification can be visualized as a flowchart:
1. Sample Preparation
Isolate cells from the tissue of interest.
2. Cell Isolation and Single-cell capture
Isolate individual cells using methods like FACS or microfluidics.
3. RNA extraction and library preparation
It’s amazing what scientists are accomplishing – building a complete catalogue of every cell type in the human body! It makes you think about the incredible complexity of life, and how much we still have to learn. This level of detailed understanding, though, could easily be hampered by political ideologies, like the ones discussed in this article, socialism is inherently evil says justin haskins of the heartland institute , which could impact funding and research priorities.
Ultimately, though, the quest to understand our cellular makeup continues, pushing the boundaries of medical knowledge.
Extract RNA from each cell, convert it to cDNA, and prepare it for sequencing.
4. Sequencing
It’s mind-blowing that scientists are building a complete catalogue of every cell type in the human body – a monumental undertaking! This kind of massive data collection, though, makes me think about the broader implications of large-scale projects and how it relates to individual liberties; it’s interesting to consider this in light of the current political climate, where, as reported by americans want less government , leading to questions about data privacy and control.
Ultimately, the cell catalogue could revolutionize medicine, but the path to that future needs careful consideration of these wider societal concerns.
Sequence the cDNA to determine the expression levels of each gene in each cell.
5. Data processing and quality control
Filter low-quality data and normalize the gene expression data.
It’s amazing what scientists are accomplishing these days; they’re building a complete catalogue of every cell type in the human body! This incredible undertaking could revolutionize medicine, but I worry about the economic implications. I read this alarming article today about unemployment seen climbing much higher than the Fed expects as it fights inflation, according to Deutsche Bank , which could impact funding for crucial research like this cell atlas project.
Hopefully, we can prioritize both scientific advancement and economic stability. Ultimately, a deeper understanding of our cells is key to future health breakthroughs.
6. Dimensionality reduction
Reduce the dimensionality of the data using techniques like PCA or t-SNE.
7. Clustering
Group cells with similar gene expression profiles into clusters.
8. Marker gene identification
Identify genes that are differentially expressed between clusters.
9. Cell type annotation
Assign cell type identities to clusters based on marker gene expression and prior knowledge.1
0. Validation
Validate the cell type assignments using independent methods such as immunohistochemistry or functional assays.
Applications of the Cell Catalogue
The creation of a comprehensive cell catalogue, such as the Human Cell Atlas, represents a monumental leap forward in biological understanding. This detailed inventory of all cell types in the human body holds immense potential for revolutionizing various aspects of medicine and biological research, far beyond simply classifying cells. Its applications span disease diagnosis, drug discovery, and the future of personalized medicine.The sheer detail and scope of the cell catalogue offer unprecedented opportunities.
By providing a baseline of normal cellular composition and function, it allows researchers to identify deviations associated with disease with greater precision than ever before. This opens doors to earlier and more accurate diagnosis, leading to improved treatment outcomes.
Disease Diagnosis and Treatment
The cell catalogue facilitates the identification of disease-specific cellular signatures. For instance, by comparing the cellular composition of a tumor biopsy to the catalogue’s normal cell profiles, researchers can identify unique cell types or aberrant cellular activities characteristic of the cancer. This information can be used to refine diagnostic tests, predict disease progression, and tailor treatment strategies. Imagine identifying a specific rare immune cell subset associated with the rapid progression of a particular leukemia; this would allow for earlier and more targeted intervention.
Similarly, identifying specific inflammatory cell populations in a patient’s blood sample could help diagnose autoimmune diseases much earlier than currently possible.
Drug Discovery and Development
The cell catalogue provides a powerful resource for drug discovery and development by enabling researchers to identify novel drug targets and predict drug efficacy and toxicity. For example, researchers can screen drugs against specific cell types identified in the catalogue to identify those most effective against a particular disease. This targeted approach reduces the time and cost associated with traditional drug discovery methods and increases the likelihood of success.
The catalogue also allows for the identification of cells that may be particularly vulnerable to drug-induced toxicity, allowing for safer drug development and minimizing side effects. Consider the development of a drug targeting a specific cancer cell type identified only through the detailed analysis offered by the catalogue. This precise targeting would minimize damage to healthy cells, significantly improving treatment outcomes.
Personalized Medicine
The cell catalogue is fundamental to the advancement of personalized medicine. By characterizing the cellular composition of an individual’s tissue, it is possible to develop tailored treatment strategies based on their unique cellular profile. This allows for the prediction of individual responses to different treatments and the optimization of therapeutic interventions. For instance, a patient’s response to immunotherapy could be predicted based on the abundance of specific immune cells in their tumor microenvironment, identified through comparison with the cell catalogue.
This would allow clinicians to select the most effective immunotherapy regimen for each patient, maximizing the chance of success and minimizing unnecessary side effects.
Future Applications of the Human Cell Atlas
The potential applications of the Human Cell Atlas extend far beyond the current scope of its development. The detailed information provided by this resource will undoubtedly fuel innovation in many areas of biological research and medicine.
- Understanding Development and Aging: Tracking cellular changes throughout the lifespan can provide insights into the mechanisms of aging and age-related diseases.
- Regenerative Medicine: The catalogue can guide the development of new cell-based therapies and tissue engineering strategies.
- Infectious Disease Research: Understanding how pathogens interact with different cell types can lead to the development of novel antiviral and antibacterial therapies.
- Environmental Health: Studying the effects of environmental factors on cellular composition can help assess the impact of pollution and other environmental hazards on human health.
- Precision Diagnostics: Development of more sensitive and specific diagnostic tests based on unique cellular signatures of diseases.
Challenges and Limitations
Creating a complete catalogue of every cell type in the human body is a monumental undertaking, fraught with significant challenges. The sheer complexity of the human body, with its trillions of cells exhibiting diverse states and functions across various tissues and organs, presents a formidable hurdle. Further complicating matters are the limitations of current technologies and the inherent difficulties in standardizing data across different research groups.
Technological Limitations in Cell Identification
Current technologies, while rapidly advancing, still possess limitations in accurately identifying and classifying all cell types. Techniques like single-cell RNA sequencing (scRNA-seq) provide valuable transcriptomic data, allowing researchers to profile gene expression patterns in individual cells. However, scRNA-seq data can be noisy, and interpreting the subtle differences between cell types based solely on gene expression can be challenging. Furthermore, scRNA-seq doesn’t capture the full picture of cellular identity; it omits information about protein expression, epigenetic modifications, and the cell’s spatial context within a tissue.
Other methods like mass cytometry offer high-throughput analysis of protein expression, but are limited in the number of markers that can be simultaneously measured. The integration of data from multiple techniques is crucial but presents significant computational and analytical challenges. For instance, integrating scRNA-seq data with spatial transcriptomics data to understand the location and function of cells within their tissue microenvironment requires sophisticated computational tools and algorithms.
Data Standardization Challenges Across Research Groups
Inconsistencies in experimental protocols, data processing pipelines, and annotation strategies across different research groups pose a major obstacle to building a comprehensive and reliable cell catalogue. Variations in sample preparation, cell isolation methods, and sequencing platforms can introduce significant technical biases. Different research groups may also use different ontologies and nomenclature systems for classifying cell types, making it difficult to compare and integrate data from multiple studies.
The lack of standardized protocols and data formats hinders the ability to create a unified and universally accessible cell catalogue. For example, one group might define a specific cell type based on the expression of a particular gene, while another group might use a different set of markers, leading to discrepancies in cell type classification.
Potential Biases in the Data and Mitigation Strategies
Several biases can affect the accuracy and representativeness of the cell catalogue. Sampling bias, for example, can occur if the cell populations sampled are not representative of the entire body. This is particularly relevant for cells that are rare or located in difficult-to-access tissues. Another potential bias is batch effects, which can arise from variations in experimental conditions across different batches of samples.
To mitigate these biases, researchers need to employ rigorous experimental designs, including careful sample collection and processing, standardized protocols, and appropriate statistical methods for data normalization and analysis. The use of large, diverse cohorts of individuals is also crucial for capturing the heterogeneity of cell types across different individuals and populations. Furthermore, blind analysis and independent validation of findings can help reduce subjective biases in data interpretation.
For instance, the development of standardized pipelines for data analysis and integration across different platforms can help to minimize batch effects and ensure data comparability.
Data Visualization and Accessibility
Creating a comprehensive cell catalogue is only half the battle; making this invaluable resource readily accessible and understandable to the global research community is equally crucial. Effective data visualization is key to unlocking the catalogue’s potential, allowing researchers to explore complex relationships between cell types and their functions. This section details strategies for visualizing the data, ensuring accessibility, and maintaining data integrity.
Visual Representation of the Cell Catalogue
For a scientific publication, the cell catalogue could be visually represented as an interactive, multi-layered network graph. The nodes would represent individual cell types, with size proportional to the abundance of that cell type in the body. The edges connecting the nodes would represent known functional relationships or developmental pathways between cell types, with edge thickness reflecting the strength of the relationship.
Different colors could be used to categorize cells based on tissue origin, lineage, or functional characteristics. The entire graph would be navigable, allowing users to zoom in on specific regions of interest or filter the display based on various parameters. This interactive approach would allow for exploration of complex cellular relationships and facilitate hypothesis generation. Tooltips hovering over nodes and edges could provide detailed information about each cell type and its connections.
Methods for Data Accessibility
Making the cell catalogue accessible requires a multi-pronged approach. First, the data should be stored in a standardized, open-access database using a well-documented and widely accepted format, such as HDF5 or a relational database like PostgreSQL. This ensures compatibility with various bioinformatics tools. Second, a user-friendly web interface should be developed, allowing researchers to query the database using various search parameters (e.g., cell type name, tissue origin, gene expression profile).
The interface should also incorporate the interactive network graph described above, providing a visual representation of the data. Finally, the data should be made available through established data repositories like NCBI GEO or the European Nucleotide Archive, ensuring long-term preservation and discoverability. Regular updates and version control will be vital to maintain data currency and track changes.
Data Integrity and Security, Scientists are building a catalogue of every type of cell in our bodies
Maintaining data integrity and security is paramount. A robust data validation pipeline should be implemented to ensure data accuracy and consistency. This includes automated checks for inconsistencies, outliers, and errors during data entry and processing. Version control systems should be used to track changes made to the database, allowing for easy rollback in case of errors. Data security measures should be implemented to protect the data from unauthorized access and modification.
This includes secure storage solutions, access control mechanisms, and regular security audits. Data encryption should be employed both in transit and at rest. A detailed data usage policy should be established, outlining acceptable uses of the data and the responsibilities of users.
Data Visualization Techniques and Suitability
The choice of data visualization technique depends heavily on the type of cell data being presented. Below is a table outlining some common techniques and their suitability:
| Visualization Technique | Data Type | Advantages | Disadvantages |
|---|---|---|---|
| Scatter Plot | Gene expression levels (two genes) | Simple, effective for showing correlations | Limited to two dimensions; can be cluttered with large datasets |
| Heatmap | Gene expression levels (many genes) | Shows patterns across many samples and genes | Can be difficult to interpret with many genes or samples |
| Network Graph | Cell-cell interactions, pathways | Visualizes complex relationships; interactive exploration possible | Can be complex to create and interpret for very large networks |
| PCA Plot | High-dimensional data (e.g., gene expression profiles) | Reduces dimensionality, reveals major sources of variation | Loss of information due to dimensionality reduction; interpretation can be challenging |
Impact on Scientific Understanding: Scientists Are Building A Catalogue Of Every Type Of Cell In Our Bodies
The Human Cell Atlas (HCA), a comprehensive catalogue of all human cell types, promises a transformative shift in our understanding of human biology and medicine. Its impact extends far beyond simply creating a detailed inventory; it provides a foundational resource for unprecedented advancements across numerous fields. By offering a standardized, detailed map of the cellular landscape, the HCA empowers researchers to investigate biological processes with an accuracy and depth previously unimaginable.The catalogue’s potential to revolutionize our approach to disease research is immense.
Understanding the cellular composition of healthy tissues allows for precise identification of the cellular and molecular changes that occur during disease development. This granular level of detail enables the development of more targeted therapies and diagnostic tools. For example, identifying specific immune cell subsets involved in autoimmune diseases like rheumatoid arthritis could lead to the development of drugs that specifically target these cells, minimizing side effects while maximizing therapeutic efficacy.
Revolutionizing Disease Research
The HCA provides a framework for understanding disease at a cellular level. By comparing the cellular composition of healthy tissues with diseased tissues, researchers can pinpoint the specific cell types and molecular pathways involved in disease pathogenesis. This knowledge is crucial for developing more effective diagnostic tools and therapies. For instance, cancer research will benefit significantly; the HCA can help identify unique cellular signatures of different cancer types, leading to more precise diagnoses and personalized treatments.
The ability to track the evolution of cancer cells over time, from initial mutation to metastasis, opens avenues for early detection and intervention strategies. Furthermore, the identification of specific cell types involved in drug resistance could inform the development of novel treatment strategies to overcome this major challenge in cancer therapy.
Advancing Understanding of Development and Aging
The HCA offers a powerful tool for studying human development from conception to adulthood, providing insights into the complex cellular processes that shape our bodies. By tracking changes in cellular composition during development, researchers can identify critical stages and potential points of vulnerability. Similarly, the HCA facilitates the investigation of age-related changes at the cellular level, allowing for a deeper understanding of the aging process and the development of age-related diseases.
For example, studying the changes in the composition of the immune system during aging could reveal new targets for interventions to improve immune function and prevent age-related decline. Observing the cellular changes in the brain during aging may shed light on the mechanisms underlying neurodegenerative diseases like Alzheimer’s and Parkinson’s.
Improving Diagnostics and Treatments
The HCA’s impact on diagnostics and treatments is multifaceted. The detailed cellular maps allow for the development of more precise diagnostic tests, enabling earlier and more accurate disease detection. For example, identifying specific biomarkers expressed by diseased cells could lead to the development of liquid biopsies, which offer a less invasive alternative to traditional tissue biopsies. Furthermore, the HCA can facilitate the development of personalized medicine approaches, tailoring treatments to the specific cellular characteristics of an individual’s disease.
This personalized approach could significantly improve treatment efficacy and reduce side effects. The identification of novel drug targets, based on the unique cellular signatures of different diseases, opens avenues for the development of more effective and targeted therapies. For instance, identifying specific cell surface receptors expressed on cancer cells could lead to the development of targeted therapies that selectively eliminate cancer cells while sparing healthy cells.
The creation of a complete cell catalogue represents a paradigm shift in our understanding of human biology. The Human Cell Atlas project is not just building a database; it’s constructing a foundation for future medical advancements. From personalized medicine tailored to an individual’s unique cellular makeup to the development of revolutionary new therapies, the implications are far-reaching and incredibly exciting.
While challenges remain, the progress made so far is nothing short of inspiring, and the future potential of this project is truly breathtaking. It’s a journey of discovery that promises to reshape healthcare as we know it.

